A ReMAP scraping script
Introduction
In this short document we use the XML package to obtain and parse an HTML table from REMAP. This table contains an overview over cell-lines and transcription-factor (TF) binding sites measured in these cell-types. We further create an overview on the number of TFs per cell-types, generating a plot which shows the number of TFs, accumulated over all cell-types. The order is such that we start with the cell-types having the most TFs available and proceed with the one adding most new TFs and so forth.
REMAP
REMAP is a huge resource which collects TF binding sites (TFBS) for numerous TFs and for hundreds of cell-types. These TFBS are ChIP-seq based, an experimental protocol which enables a genome-wide readout of DNA sites which are bound by specific transcription factors. The current version (2018) contains over 80 million TFBS for 485 TFs identified in 346 cell-types.
Implementation
Data processing
First, we load needed libraries and read in the table. We could optionally save the table as a TSV to disc.
NOTE: We load the cowplot package only to get a nice and lightweight default theme for ggplot set up. Also, I can really recommend the package for publication ready figures.
library(tidyverse)
library(cowplot)
library(XML)
# read and extract HTML table
remap <- readHTMLTable("http://tagc.univ-mrs.fr/remap/index.php?page=ct")[[1]] %>%
as_tibble(.name_repair = "universal")
remap## # A tibble: 346 x 6
## Cell.Type Number.of.Publi~ Number.of.Encod~ Name BTO.ID
## <fct> <fct> <fct> <fct> <fct>
## 1 MCF7 190 71 MCF-~ BTO:0~
## 2 LNCAP 123 2 LNCa~ BTO:0~
## 3 VCAP 81 0 VCaP~ BTO:0~
## 4 ESC 69 76 embr~ BTO:0~
## 5 HELA 48 72 HeLa~ BTO:0~
## 6 T47D 48 9 T-47~ BTO:0~
## 7 U2OS 41 2 U2-O~ BTO:0~
## 8 HCT166 33 23 HCT-~ BTO:0~
## 9 HUVEC 33 0 HUVE~ BTO:0~
## 10 MM1S 33 0 MM1-~ BTO:0~
## # ... with 336 more rows, and 1 more variable: Transcription.Factor <fct># Optional: write obtained data to disc
write_tsv(remap, "../downloads/remap_celltype_tfs.tsv")This was easy enough, we got the table and transformed it into a tibble, getting nicer column names on the way. You can download the transformed table from here.
NOTE: the readHTMLTable actually returns a list of results it finds. We instantly subset the list and only retrieve the first element, which, in our case, is the main table.
Since all TFs are gathered in a single cell for each cell-type, in the next step we separate the table by the TFs in each row, using the very convenient separate_rows() method. This method expands each row into multiple rows, based on the values of a string split generated from the values of a specific column. For us, this yields for each cell-type individual rows for each available TF.
# separate
remap_sep <- remap %>%
separate_rows(Transcription.Factor)
# count TFs per cell type
remap_counts <- remap_sep %>% group_by(Cell.Type) %>%
summarize(count = n()) %>%
arrange(desc(count))
remap_counts## # A tibble: 346 x 2
## Cell.Type count
## <fct> <int>
## 1 K562 179
## 2 GM12878 110
## 3 HEPG2 103
## 4 ESC 88
## 5 HELA 85
## 6 MCF7 84
## 7 A549 50
## 8 HCT166 33
## 9 SKNSH 28
## 10 LNCAP 25
## # ... with 336 more rowsWe can see that the cell-type with the most TFs measured is the K562 (used in ENCODE), closely followed by the GM12878 LCL cell-line. In fact, most of the top cell-types were used in ENCODE.
Finally, we generate the cumulative numbers we want to plot in the end. Since we always want to add only the cell-line contributing most TFs in each step, we do this manually and evaluate the overlap of the TF lists on the way. This might be a somewhat crude implementation, but it does the job. If you know a shortcut for achieving this, let me know!
# list of cell types to process
cell_types <- as.character(remap_counts$Cell.Type)
# get all TFs available for the first cell-type (which has
# the maximum number of TFs since we use the sorted list)
tfs <- remap_sep %>% filter(Cell.Type == cell_types[1]) %>%
select(Transcription.Factor) %>%
unlist(use.names=F)
# prepare result df
df <- data.frame(cell_type=cell_types[1],
number_contr_tfs=length(tfs),
contr_tfs=paste0(tfs, collapse=","),
stringsAsFactors = F)
# remove the first cell type, iterate over all others
# as long as we've got some still left unprocessed
cell_types <- cell_types[-1]
while(length(cell_types) > 0) {
max_contr <- -1
max_contr_ct <- NA_character_
max_contr_tfs <- NA_character_
# check each remaining cell_type for max contribution
for(i in 1:length(cell_types)) {
ct_tfs <- filter(remap_sep, Cell.Type == cell_types[i]) %>%
select(Transcription.Factor) %>%
unlist(use.names=F)
# how many are not yet in the list of all tfs?
new_tfs <- setdiff(ct_tfs, tfs)
nnew_tfs <- length(new_tfs)
if(nnew_tfs > max_contr) {
max_contr <- nnew_tfs
max_contr_tfs <- new_tfs
max_contr_ct <- cell_types[i]
}
}
# in that case, we didn't have any new TFs..
if(max_contr == 0) {
# add all remaining cell-types to the df
remaining <- rbind(data.frame(cell_type=cell_types,
number_contr_tfs=rep(0, length(cell_types)),
contr_tfs=rep(NA, length(cell_types)),
stringsAsFactors = F))
df <- rbind(df, remaining)
break
}
# remember the most contributing cell type
row <- c(max_contr_ct, max_contr, paste0(max_contr_tfs, collapse=","))
tfs <- c(tfs, max_contr_tfs)
df <- rbind.data.frame(df, row)
cell_types <- setdiff(cell_types, max_contr_ct)
}Plotting
Now we can use the accumulated TF contributions for plotting with ggplot. We filter for contributions > 0 and calculate the cumulative sum for the y-axis. We further add the cell type labels to each data point using the geom_text() ggplot layer.
df_sub <- subset(df, number_contr_tfs > 0)
ggplot(df_sub, aes(x=1:nrow(df_sub), y=cumsum(number_contr_tfs))) +
geom_line() +
geom_point(size=2) +
geom_text(aes(label=cell_type), vjust=0, hjust=-0.5, check_overlap = F, angle=-25) +
scale_y_continuous(limits=c(0,500)) +
labs(title="Cumulative sum of number of new TFs contributed by each cell-type",
subtitle = "Sorted by total contribution",
y="Cumulative sum of TF contributions",
x="Cell type")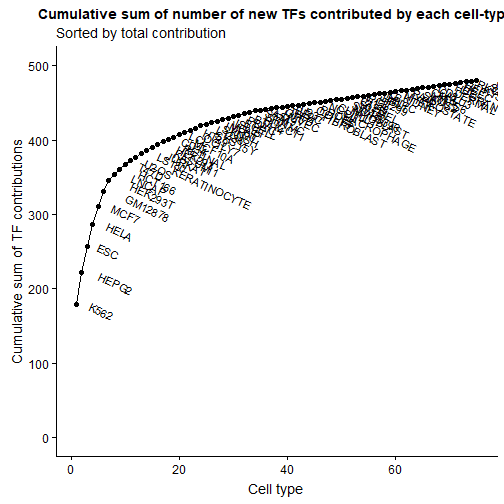
Summary
That’s all! This was a quick (not necessarily dirty) way of extracting a HTML table from a website and generating a brief overview.
Until then, farewell!
Session Info
## R version 3.5.1 (2018-07-02)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 10 x64 (build 17763)
##
## Matrix products: default
##
## locale:
## [1] LC_COLLATE=English_United States.1252
## [2] LC_CTYPE=English_United States.1252
## [3] LC_MONETARY=English_United States.1252
## [4] LC_NUMERIC=C
## [5] LC_TIME=English_United States.1252
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] XML_3.98-1.20 cowplot_0.9.4 forcats_0.4.0 stringr_1.4.0
## [5] dplyr_0.8.0.1 purrr_0.3.2 readr_1.3.1 tidyr_0.8.3
## [9] tibble_2.1.1 ggplot2_3.1.1 tidyverse_1.2.1 knitr_1.22
##
## loaded via a namespace (and not attached):
## [1] Rcpp_1.0.1 highr_0.8 cellranger_1.1.0 pillar_1.4.1
## [5] compiler_3.5.1 plyr_1.8.4 tools_3.5.1 zeallot_0.1.0
## [9] jsonlite_1.6 lubridate_1.7.4 evaluate_0.13 nlme_3.1-137
## [13] gtable_0.3.0 lattice_0.20-35 pkgconfig_2.0.2 rlang_0.3.4
## [17] cli_1.1.0 rstudioapi_0.10 haven_2.1.0 xfun_0.6
## [21] withr_2.1.2 xml2_1.2.0 httr_1.4.0 vctrs_0.1.0
## [25] generics_0.0.2 hms_0.4.2 grid_3.5.1 tidyselect_0.2.5
## [29] glue_1.3.1 R6_2.4.0 fansi_0.4.0 readxl_1.3.1
## [33] modelr_0.1.4 magrittr_1.5 backports_1.1.4 scales_1.0.0
## [37] rvest_0.3.3 assertthat_0.2.1 colorspace_1.4-1 labeling_0.3
## [41] utf8_1.1.4 stringi_1.4.3 lazyeval_0.2.2 munsell_0.5.0
## [45] broom_0.5.2 crayon_1.3.4
Comments