Example of how to create a genome-wide genomic pairs plot
Introduction
In this post, I want to give an example of how pairs of genomic entities, such as genotypes and associated genes (QTLs), can be plotted in a genome-wide scale using tidyverse/ggplot2 and the GenomicRanges package in R. We will use publicly available trans-eQTL data (see below) and will create a single overview matrix showing the frequencies of QTLs in these data. To this end, we will bin/tile all standard chromosomes of the human reference genome (GRCh37/hg19) into bins of 10MB, overlap these bins with our QTL results and then plot them using a simple tile plot available in ggplot2. In addition, we’ll add margin plots to the main matrix-like plot, showing the row and column summary counts over all associations.
Data
Expression quantitative trait loci (eQTL) are, in essence, statistical associations between genetic variants/genotypes (e.g. single nucleotide polymorphisms, SNPs) and the expression of genes measured ideally in a large number of samples to obtain sufficient statistical power. In other words, eQTL are a way of pinpointing DNA sequence variants which, most likely, have a functional impact in the many complex mechanisms taking place in the studied organism. Generally, eQTL can be classified in cis- and trans-eQTL, cis meaning that the SNP and the respective gene reside on the same chromsome and trans meaning that the two entities are located on different chromosomes. For our example, we will focus on trans-eQTL only. We downloaded all significant trans-eQTL data identified by the eQTLgen consortium and use those to generate a nice overview on trans-eQTLs identified in whole blood data.
Hands-on
Data preparation
As a first step, we load the eQTLgen trans-eQTLs and create GenomicRanges objects, i.e. objects to easily handle genomic position annotations. We also filter for ‘real’ trans eQTL (a matter of definition), in our case this means that SNP and Gene are on distinct chromosomes.
# load eQTL data
#eqtl <- read_tsv("https://molgenis26.gcc.rug.nl/downloads/eqtlgen/trans-eqtl/trans-eQTL_significant_20181017.txt.gz")
eqtl <- read_tsv("d:/eQTLgen/trans-eQTL_significant_20181017.txt.gz") %>%
filter(SNPChr != GeneChr)
# get the number of associations per SNP
ntrans_per_snp <- group_by(eqtl, SNP) %>% summarise(ntrans=n())
# filter hotspots, i.e. retain only SNPs with at least 5 trans associations
eqtl <- mutate(eqtl, ntrans = ntrans_per_snp[match(SNP, ntrans_per_snp$SNP),]$ntrans) %>%
filter(ntrans >= 5)
# get the hg19 chromosome definitions
library(BSgenome.Hsapiens.UCSC.hg19)
hg19info <- seqinfo(BSgenome.Hsapiens.UCSC.hg19)
# create the ranges objects
eqtl_ranges <- with(
eqtl,
GRanges(
paste0("chr", SNPChr),
IRanges(SNPPos, width = 1),
name = SNP,
trans_associations = ntrans,
seqinfo = hg19info
)
)
trans_genes <- with(eqtl,
GRanges(
paste0("chr", GeneChr),
IRanges(GenePos, width = 2),
name = GeneSymbol,
seqinfo = hg19info
))Ok, nice! We now got all our data loaded and have them neatly available as GenomicRanges objects. We are looking at SNPs which are associated with a total of 5455 genes ( total associations). Now, since we want to do a genome-wide plot, we need to get some information on the size of the chromosomes etc. in order to be able to indicate chromosome boundaries. For that, we get the hg19 genome annotation and extract the sequence lengths for chromosomes 1-22. We further tile our genome information into tiles (or bins) of size 10MB. These tiles will be used later on to map the individual genomic positions from the SNPs and genes to the respective position in the plot.
# get chromoeome lengths
chrs = paste0("chr", 1:22)
chrlen <- seqlengths(hg19info)
chrlen <- chrlen[paste0("chr", 1:22)]
# tile the genome
genome_bins <-
tileGenome(chrlen,
tilewidth = 1e6,
cut.last.tile.in.chrom = T)
# define breaks (used to get a nice, scaled
# grid visualization)
breaks <- table(seqnames(genome_bins))
for (i in 2:length(breaks)) {
breaks[i] <- breaks[i - 1] + breaks[i]
}With the above steps done, we can now almost start creating the plot. Last remaining things to do is to map our eQTL entities to the respective genome bins and to create the summary counts for the x and y (row and column) margins. Let’s do that now:
# get overlaps
bin_overlaps_eqtl <- findOverlaps(eqtl_ranges, genome_bins)
bin_overlaps_genes <- findOverlaps(trans_genes, genome_bins)
# to be save, we only retain pairs where both entities could be maped
# to chromosomes 1:22
mappable_pairs <-
intersect(queryHits(bin_overlaps_eqtl), queryHits(bin_overlaps_genes))
x_bin <-
subjectHits(bin_overlaps_eqtl)[queryHits(bin_overlaps_eqtl) %in% mappable_pairs]
y_bin <-
subjectHits(bin_overlaps_genes)[queryHits(bin_overlaps_genes) %in% mappable_pairs]
# create a data.frame for plotting with ggplot2
pairs_binned <- cbind.data.frame(x_bin, y_bin)
pairs_binned <-
pairs_binned[order(pairs_binned$x_bin, pairs_binned$y_bin),]
# the margins, i.e. the individual totals for the x and y bins
x_margin <- group_by(pairs_binned, x_bin) %>% summarise(count = n())
y_margin <- group_by(pairs_binned, y_bin) %>% summarise(count = n())Nice, now everything is prepared and we can start with the actual plotting!
The actual plotting
Let’s keep it brief, I added some comments to the code (as I’m supposed to do anyway) so you can figure out what is happening, but I kept it in a single code block so you can just copy, paste and adjust if you want.
# Since we use cowplot, we do not perform any theme adjustments just yet
# get some nice qualitative colors
cols <- brewer.pal("Set2", n=3)
color <- cols[1]
# create the plot for the x-axis margin, single points
xmp <- ggplot(x_margin) +
geom_point(aes(x=x_bin, y=count), color=color, shape=23) +
scale_x_continuous(expand = c(0.01, 0.01), breaks = as.vector(breaks), labels = NULL) +
xlab("") +
background_grid(major = "xy")
ymp <- ggplot(y_margin) +
geom_point(aes(x=y_bin, y=count), color=color, shape=23) +
coord_flip() + scale_x_continuous(
expand = c(0.01, 0.01),
breaks = as.vector(breaks),
labels = NULL) +
xlab("") +
theme(axis.text.x = element_text(angle=90,
vjust=0.5,
hjust=1)) +
background_grid(major = "xy")
# the main matrix plot showing the eQTL information
g <-
ggplot(pairs_binned) +
geom_tile(fill=color, width=10,
aes(x = x_bin, y = y_bin)) +
theme(
text = element_text(size = 11),
legend.text = element_text(size = 8),
axis.text.x = element_text(vjust = 0.5, angle = 90),
legend.title = element_text(size = 10),
plot.margin = margin(0,0.1,0,0, "cm")) +
xlab("SNPs") + ylab("Genes") +
scale_x_continuous(
expand = c(0.01, 0.01),
breaks = as.vector(breaks),
labels = names(breaks),
limits = c(1,length(genome_bins))
) +
scale_y_continuous(
expand = c(0.01, 0.01),
breaks = as.vector(breaks),
labels = names(breaks)
) + background_grid(major = "xy")
# now we got the individual plots, arrange them in a single
# large plot using grid.arrange
wr <- c(85,15)
hr <- c(15,85)
side_margins <- margin(0.1,0.7,0.1,-0.9, unit = "lines")
top_margins <- margin(0.7, 0,-0.6, 0.1, unit = "lines")
# eqtl matrix plot, slightly modify needed parts
xmp <- xmp + theme(plot.margin = top_margins)
ymp <- ymp + theme(plot.margin = side_margins)
# now we perform the actual plot,
# in this case all gets a little bit crowded, I try to adjust this at a later time
# but you get the idea
plot_grid(xmp, nullGrob(), g, ymp,
ncol=2, nrow=2,
rel_widths = wr, rel_heights = hr)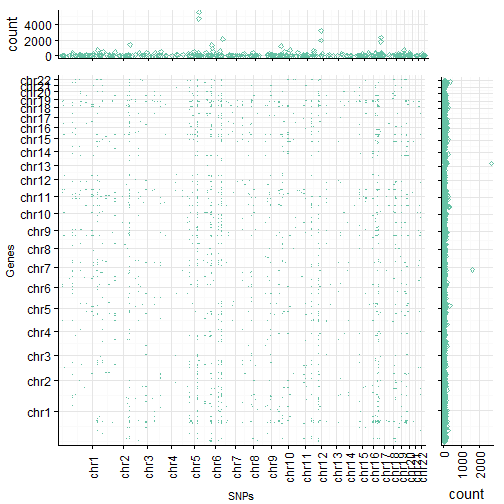
So, as you can see, we create our final plot by arranging three distinct plots in a single frame using quite some functions from different packages (e.g. plot_grid from the cowplot package, and the nullGrob() method from the grid package).
NOTE: it could happen that the axes are not perfectly aligned with the margins. the plot_grid() function provides an argument to tackle this (align and axis), but it can get tricky to try to align the axes in both dimensions!
The row and column annotations show the total number of entities falling into the respective row or column and the red dots in the main plot indicate the presence of a SNP (columns) associated with a gene (row) for the two chromosome regions. This actually looks rather good (well, good enough)! So good in fact that, we are content for now and stop tweaking the plot (I’m lazy today). Anyway, we can see that there are some SNPs or LD blocks which exhibit a huge number of trans associations as well as some regions in the genome which harbor a relatively larger number of genes which are influenced by those SNPs in trans as compared to other regions.
Conclusion
Alright, you have seen how we can get an impression of genome-wide results from an eQTL study, just by using tidyverse/ggplot2 and the GenomicRanges packages in R. Of course, there are different ways of doing this, for example you could try to do a circos plot using the ggbio package in R or you can add continuous information about the number of eQTLs per bin directly in the main matrix plot (e.g. using the fill aesthetics). Maybe I’ll extend the plot later to show how to do this, but in the meanwhile: have fun playing around with this example!
Until then, farewell!
Session Info
## R version 3.5.1 (2018-07-02)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 10 x64 (build 17763)
##
## Matrix products: default
##
## locale:
## [1] LC_COLLATE=English_United States.1252
## [2] LC_CTYPE=English_United States.1252
## [3] LC_MONETARY=English_United States.1252
## [4] LC_NUMERIC=C
## [5] LC_TIME=English_United States.1252
##
## attached base packages:
## [1] grid parallel stats4 stats graphics grDevices utils
## [8] datasets methods base
##
## other attached packages:
## [1] BSgenome.Hsapiens.UCSC.hg19_1.4.0 BSgenome_1.48.0
## [3] rtracklayer_1.40.6 Biostrings_2.48.0
## [5] XVector_0.20.0 cowplot_0.9.4
## [7] RColorBrewer_1.1-2 gridExtra_2.3
## [9] GenomicRanges_1.32.7 GenomeInfoDb_1.16.0
## [11] IRanges_2.14.12 S4Vectors_0.18.3
## [13] BiocGenerics_0.26.0 reshape2_1.4.3
## [15] forcats_0.4.0 stringr_1.4.0
## [17] dplyr_0.8.0.1 purrr_0.3.2
## [19] readr_1.3.1 tidyr_0.8.3
## [21] tibble_2.1.1 ggplot2_3.1.1
## [23] tidyverse_1.2.1 knitr_1.22
##
## loaded via a namespace (and not attached):
## [1] Rcpp_1.0.1 lubridate_1.7.4
## [3] lattice_0.20-35 Rsamtools_1.32.3
## [5] assertthat_0.2.1 R6_2.4.0
## [7] cellranger_1.1.0 plyr_1.8.4
## [9] backports_1.1.4 evaluate_0.13
## [11] highr_0.8 httr_1.4.0
## [13] pillar_1.4.1 zlibbioc_1.26.0
## [15] rlang_0.3.4 lazyeval_0.2.2
## [17] readxl_1.3.1 rstudioapi_0.10
## [19] Matrix_1.2-14 labeling_0.3
## [21] BiocParallel_1.14.2 RCurl_1.95-4.12
## [23] munsell_0.5.0 DelayedArray_0.6.6
## [25] broom_0.5.2 compiler_3.5.1
## [27] modelr_0.1.4 xfun_0.6
## [29] pkgconfig_2.0.2 tidyselect_0.2.5
## [31] SummarizedExperiment_1.10.1 GenomeInfoDbData_1.1.0
## [33] matrixStats_0.54.0 XML_3.98-1.20
## [35] crayon_1.3.4 withr_2.1.2
## [37] GenomicAlignments_1.16.0 bitops_1.0-6
## [39] nlme_3.1-137 jsonlite_1.6
## [41] gtable_0.3.0 magrittr_1.5
## [43] scales_1.0.0 cli_1.1.0
## [45] stringi_1.4.3 xml2_1.2.0
## [47] generics_0.0.2 tools_3.5.1
## [49] Biobase_2.40.0 glue_1.3.1
## [51] hms_0.4.2 colorspace_1.4-1
## [53] rvest_0.3.3 haven_2.1.0
Comments